Let’s look into the molecules involved in mitochondria’s electron transport chain and oxidative phosphorylation in more detail.
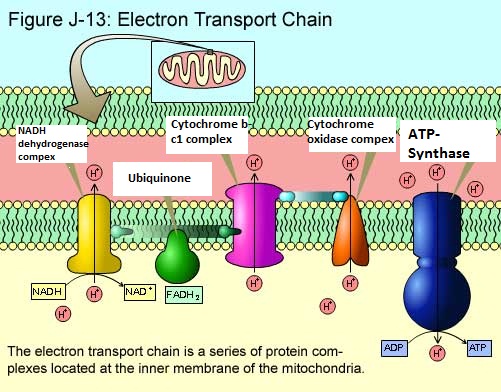
Present on the cristae (inner mitochondrial membrane) are over forty (40) proteins, fifteen (15) of which are directly involved in electron transport. Most of the proteins involved in this are grouped into three large respiratory enzyme complexes.
1- NADH dehydrogenase complex. This enzyme removes the hydrogen to make it NAD.
2- Cytochrome b-c1 complex. Second receiver of the electron.
3- Cytochrome oxidase complex. Final receiver that will donate to oxygen.
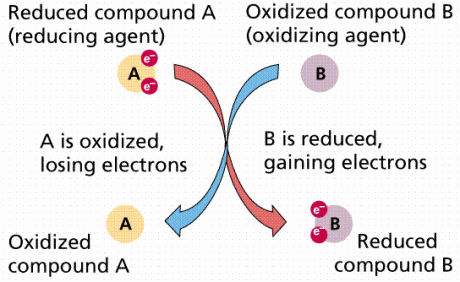
Each enzyme contains metal ions, either iron (Fe) or copper (Cu) in their heme-group (heme- iron or any readily oxidize-able metal ion) that allow passage of electrons through them. Their atomic state changes from being reduced (being more electronegative) when accepting an electron to being oxidized (being less electronegative) when losing an electron.
This redox reaction (reduction followed by oxidation) is responsible for the colour change of the protein heme-group in the enzyme complex which is why the word cytochrome (colourful) is used to describe the enzymes. For example, the iron atom changes from ferric (Fe3+) which is yellow to ferrous (Fe2+) which is blue, when it gets reduced (gains electrons to become more electronegative).
During the time that the respiratory enzyme complexes transfer the electrons in the chain, they are also in the same instance pumping protons from the inner mitochondria matrix (with the help of ubiquinone and cytochrome-c) into the intermembrane space. The way this is done is they pick up a proton from the mitochondrial matrix by attracting it to the electron (during the temporary moment when the electrons are in their possession), and then releasing proton into the intermembrane space as the electron is passed on to the next respiratory enzyme complex in the chain.

Allosteric changes creates sequential changes in protein configuration that causes the H+ to enter. Click on image for credit.
The attraction occurs in the form of allosteric (cooperative binding) changes, because the temporary accommodation of the electron within the respiratory enzyme complex creates sequential changes in protein configuration that causes the H+ to enter. Once the electron gets transferred, a similar change in the protein configuration of the enzyme causes the H+ to be released on the other side in the intermembrane space. Think of a newly opened hotel trying to gain guests. It will put out an advertisement of discounted buffet at its dining hall. The people who attend the buffet will then be shown the hotel rooms. Thus, coupling the attraction to the buffet in order to gain prospective clients is similar to coupling the attraction to the electron to attract and pump protons out. The hotel is the enzyme complex, the buffet is the electron, the people attracted to the buffet are the proton, and the hotel rooms (being the intended destination) is the intermembrane space.
Moreover, the active pumping of protons has two major consequences:
- It generates a gradient of proton concentration (a pH gradient) across the inner mitochondrial membrane, with the pH about 1 unit higher in the matrix (around pH 8) than in the intermembrane space (close to pH 7).
- It generates a membrane potential across the inner mitochondrial membrane, with the inside (matrix side) negative and outside positive as a result of net outflow of protons.
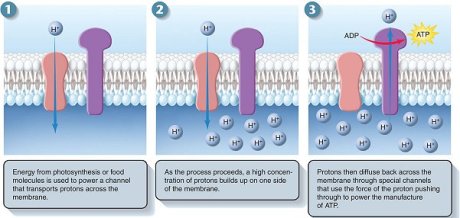
This steep electrochemical proton gradient makes it energetically very favourable for H+ to flow back into the mitochondrial matrix. About 3 protons need to pass through the synthase to make one molecule of ATP (100 molecules are made per second).
ATP synthase is a large, multisubunit protein with an enzymatic portion that looks like a lollipop head and faces inwards and attached, through a thinner multisubunit “stalk”, to a transmembrane proton carrier. The movement of protons down their gradient causes the stalk to spin rapidly within the head inducing it to make ATP.

ATP Synthesis. Credit is due, unfortunately lost the original link. Please inform me to relink if possible.
Btw, the synthesis of ATP is not the only process driven by the gradient, charged molecules (like pyruvate). ADP and Pi are pumped into the matrix from the cytosol while ATP must be moved in the opposite direction. This occurs simultaneously in other carrier proteins present in the cristae which binds these molecules together and couples their transport to the energetically favourable flow of H+ into the mitochondrial matrix (remember cotransport?).